New Method Reveals Mechanisms of the Immune System
Monday, 23.05.2022
New Method Reveals Mechanisms of the Immune System
The Uetrecht group (Uni Siegen/ LIV), at the Centre for Structural Systems Biology CSSB, and collaborators including the CSSB Protein Characterisation Facility have developed the basis for a new method to predict the immune systems’ response to individual peptides. The research study, published in Communications Biology, demonstrates how a novel high-throughput screen could help identify specific peptides that bind to Major histocompatibility complex (MHC) class I molecules. According to the researchers, the efficient identification of these peptides will contribute to the development of peptide vaccines.
MHC class I molecules are cell surface proteins that alert the immune system to the existence of foreign substances and trigger an initial immune response. When MHC molecules drift by a peptide with a high affinity, it binds to the peptide and presents it to cytotoxic T cells on the cell’s surface. The T cells are able to detect foreign peptide/MHC combinations and induce apoptosis in the cell.
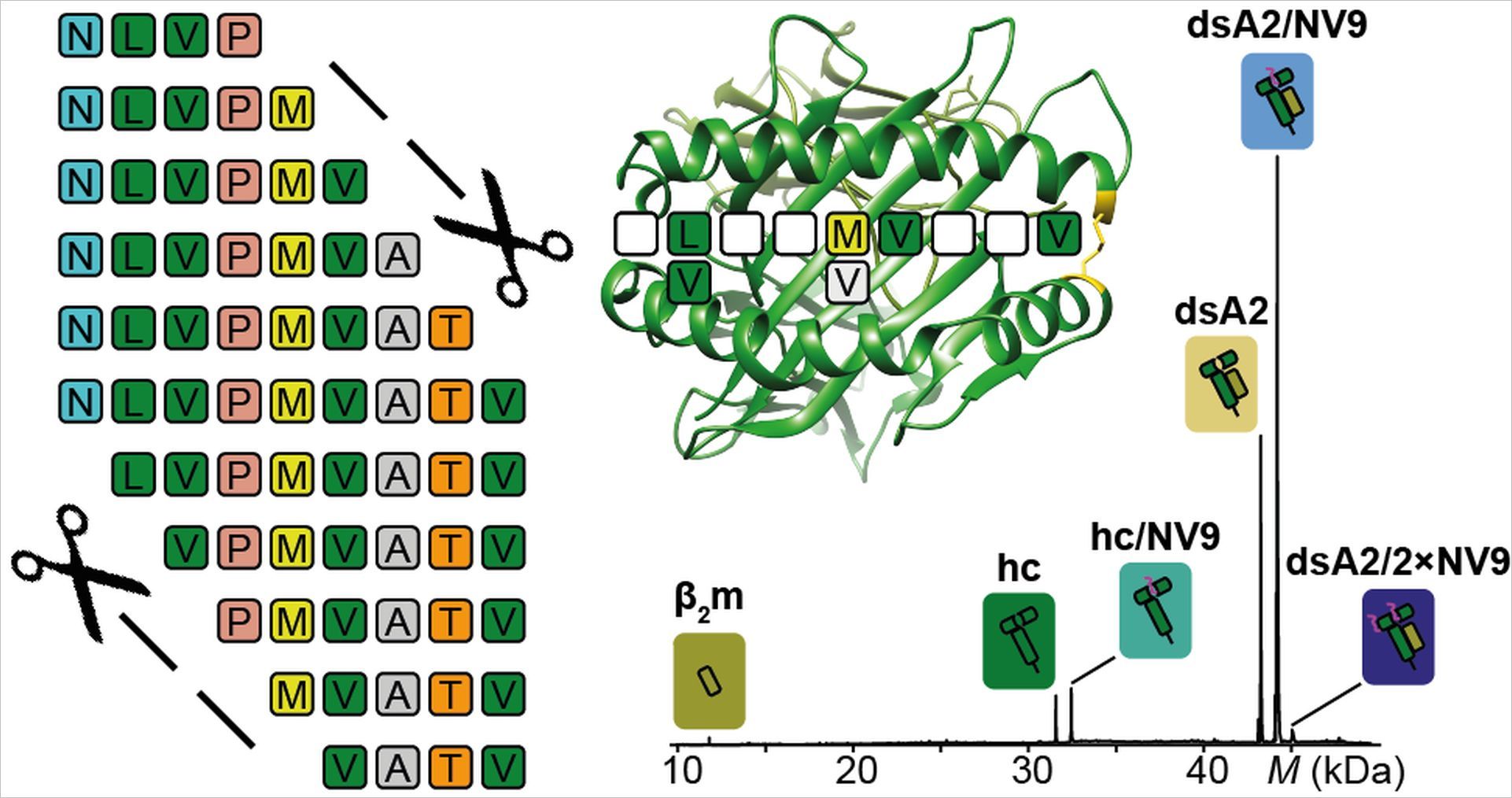
MHC molecules are complexes formed by two subunits that fall apart when no peptide is attached, therefore it is difficult to look at the structural features of the complex. In a previous paper, the researchers demonstrated a method for stabilizing an MHC molecule with a cystein bridge at the outer peptide binding pocket. These empty disulfide-stabilized class I molecules (dsMHC) bound to peptides could be examined by the researchers using native mass spectrometry (nMS), which detects all the different mass species present in a solution.
The researchers then examined MHC-peptide binding with several different peptides to determine which specific features, such as peptide length, sequence and termini, contribute to sufficient binding. “Our results were, however, not what we had expected,” explains the paper’s first author Janine-Denise Kopicki “We kept finding tiny traces of an unknown molecule other than the peptide that was not only consistently binding with the MHC molecules, but also occupying 100% of the binding pockets when no peptide was present.”
Thomas Dülcks from University of Bremen, who specializes in MS of smaller molecules, was able to identify this molecule as erucamide, a softener used in plastics, most likely from the test tubes used in the experiments. The researchers realized that erucamide’s high affinity binding could be used to determine a peptides affinity based on the fraction of erucamide’s replacement. “Our mystery molecule had suddenly given us the basis for developing a novel high-throughput peptide screen for MHC class I epitopes,” explains group leader Charlotte Uetrecht “Importantly, this new method could aid in the development of peptide vaccines which, unlike conventional vaccines and RNA vaccines, are stable and could be easily used in tropical and hot climates with limited medical infrastructure.”
Latest news about Leibniz ScienceCampus InterACt