Integrative Virology
Head: Prof. Dr. Maya Topf
The attachment of viruses to the cell, their entry, uncoating, biosynthesis, assembly, and release, all involve the formation of protein-protein interactions between the viral proteins themselves and between viral and host proteins. Thus, it is of great biomedical interest to understand the structure of complexes formed by these interactions, with the aim of designing antiviral therapies that can block them, or alternatively re-engineer viruses for use as targeted delivery vehicles. In recent years, the use of integrative, information-driven approaches for modeling structures of complexes such as those formed by viruses in cells has been increasing in popularity. The Research Department of Integrative Virology applies an integrated systems biology approach that combines computational and experimental methods specifically designed to achieve a comprehensive picture of viral infections, with specific focus on cryoEM, mass spectrometry-based proteomics and bioinformatics.
Research Topics
3D-EM Model Validation and Refinement
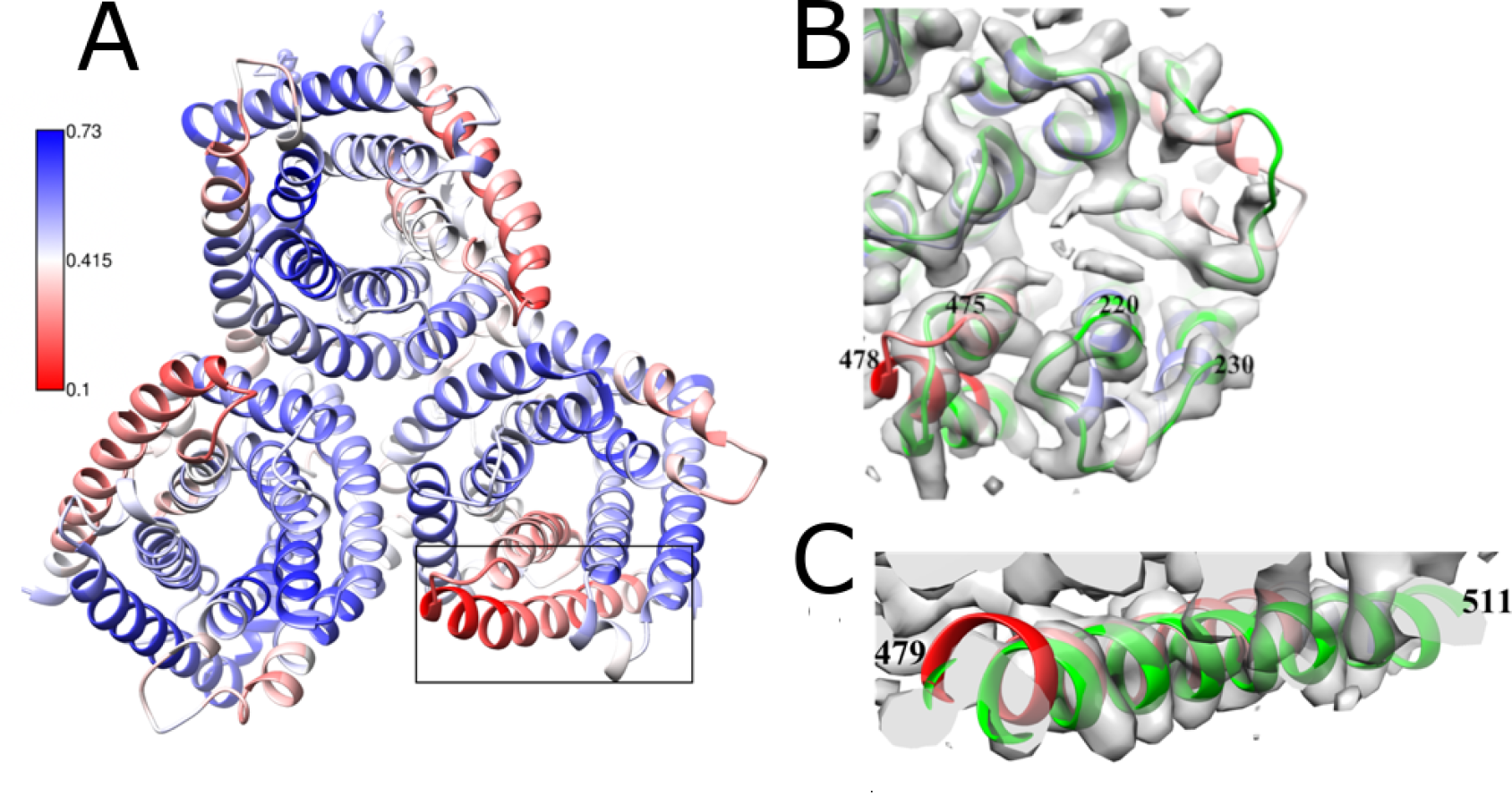
Our group works on the development of tools for fitting and assessment of atomic models in the cryo EM maps. These methods are used to characterise macromolecular assemblies from viruses. We have developed the fitting and validation software TEMPy. Recently, we assessed protein-protein interfaces in macromolecular assemblies derived by cryo EM. Using a machine learning-based metric (PI-score), we evaluated 5873 interfaces in 1053 PDB-deposited cryo EM atomistic models (including SARS-CoV-2 complexes), and in the models submitted for CASP13 cryo EM targets and the EM model challenge.
Joseph et al. Methods 2016, Cragnolini Acta D 2021, Malhotra et al. Nat. Commun. 2021
Mass Spectrometry-based Modelling
Chemical Cross-Linking Mass Spectrometry (XL-MS) is a technique which provides solvent accessible distance restraints for proteins and protein complexes in solution. The experiment produces 4 different types of peptides: cross-links, mono-links, loop-links and unmodified peptides. We developed a scoring function called Matched and Non-accessible Cross-Link (MNXL) to score models of protein structure based on cross-linking information, using both the solvent accessibility of the cross-linked residues and the distance between the two. Mono-links provide only solvent accessibility information, yet they are far more abundant in an XL-MS experiment than cross-links. We therefore also developed an approach to scoring protein models using mono-links. Our combined crosslink-monolink score is called XLMO.
Matthew Allan Bullock et al. MCP 2016; Bullock et al. 2018 Structure, Bioinformatics; Sinnott et al. Structure 2020
Protein-Protein Interaction Networks
We use protein-protein interaction (PPI) network analysis to bring new functional characterisation to virus families. We developed a computational framework for PPI network assembly, which combines both experimentally validated and computationally predicted PPI data. Using this framework, we created system-level compilations of the binary interactions among virally-encoded proteins. We applied it to three different species of human herpesviruses. Using consensus clustering approach we studied the community structure of the network data and revealed higher-order functional associations among viral proteins. Combined with primary sequence analysis of individual proteins, this study brought new functional insights on previously poorly characterised proteins.
Hernández Durán et al, PLoS Biol 2019, Hernández Durán et al mSystems 2019